

| Preprints/Archives |
|---|
| Selected papers |
| Blogs & Articles in Chinese 中文小品 |
| All publications
- Books |
Feng-Hsiang Chung, Zhen-Hua Jin, Tzu-Ting Hsu, Chueh-Lin Hsu, HC Lee
GSCMap - A Gene-Set-based version of Connectivity Map and an application using
Local Hierarchical Clustering to characterize Bioactive Compounds in terms of Biological Functional Groups
(Submitted; under journal requested revision) (PDF)
Integration of network theory with gene expression data on disease progression significantly improves prediction for human colorectal cancer biomarkers
FH Chung, HD Chen, HHC Lee and HC Lee
[ PDF]
Data processing approach for localizing bio-magnetic sources in the brain
Hung-Yi Bai, Chih-Yuan Tseng and HC Lee
ArXiv:0903.0859v1 [q-bio.QM]
[ PDF ]
Filter Out High Frequency Noise in EEG Data Using The Method of Maximum Entropy
Chih-Yuan Tseng and HC Lee
[ ArXiv:0711.2861]
[ PDF ]
Genomes: at the edge of chaos with maximum information capacity
SG Kong, HD Chen, WL Fan, J Wigger, A Torda and HC Lee
[
ArXiv:0708.1598v1 ]
[ PDF]
Numerical models of genome growth
J Wigger, SG Kong, HD Chen, WL Fan, HC Lee and A Torda
[ PDF]
Integrity of H1 helix in prion protein revealed by molecular dynamic simulations to be especially vulnerable to changes in the relative orientation of H1 and its S1 flank
Chi-Yuan Tseng, Jun-Ping Yu and HC Lee
[ ArXiv:q-bio/0603033v2
] [ PDF]
Identifying Biomagnetic Sources in the Brain by the Maximum Entropy Approach
Hung-I Pai, Chih-Yuan Tseng and HC Lee
[ ArXiv:q-bio/0508040]
[ PDF ]
Kinetics and Thermodynamics in the Folding of Trp-Cage:
Simulation by Parallel-Tempering
Jia-Lin Lo, Chun-Ping Yu and HC Lee
[ PDF]
Oligo-distance: a sequence distance determined by word frequencies
LC Hsieh, CY Tseng, LF Luo, FM Ji and HC Lee
[
JBCB preprint ]
[ PDF]
Evolutionary Tree Based on Oligonucleotide Frequencies and
Conserved Words in 16S and 18S Ribosomal RNA
LC Hsieh, CY Tseng, LF Luo, MW Jia, FM Ji and HC Lee
[PDF]
LC Hsieh, CH Chang, TY Chen, WL Fan and HC Lee
Universality in Large-Scale Structure of Complete Genomes
Transactions on Biology and Biomedicine (MABE 2004)
[ PDF]
A Universal Signature in Whole Genomes
LC Hsieh, TY Chen, CH Chang and HC Lee
[PDF] (read only)
A Universal Signature in Whole Genomes - Supporting Online Material
LC Hsieh, TY Chen, CH Chang and HC Lee
[PDF] (read only)
Universality in Large-Scale Structure of Complete Genomes
LC Hsieh, CH Chang, TY Chen, WL Fan and HC Lee [ (MABE 2004, Corfu) PDF ]
Universal Lengths in Microbial Genomes and Implication for
Early Genome Growth
CH Chang, LC Hsieh, LF Luo and HC Lee [PDF] (read only)
Quasireplicas and universal lengths of microbial genomes
LC Hsieh, LF Luo, FM Ji and HC Lee [arXiv.org/abs/physics/0309006
] [PDF] (read only)
Short Segmental Duplication: Model for Growth of
Microbial Genomes
LC Hsieh, LF Luo and HC Lee [ PDF]
Short Segmental Duplication: Parsimony in the Growth of
Microbial Genomes
LC Hsieh, LF Luo and HC Lee [PDF] (read only)
Growth of microbial genomes by
short segmental duplications
LC Hsieh, LF Luo and HC Lee [PDF] (read only)
Evidence for growth of microbial genomes by
short segmental duplications
LC Hsieh, LF Luo and HC Lee [arXiv.org/abs/physics/0302031
] [PDF]
Uptake Signal Sequences in the Genome of Haemophilus influenza
T.Y. Chen, R.J. Redfield and H.C. Lee (interim report) [PDF]
Global comparison of thirteen genomes and phylogeny
W.G. An, Z.H. Wang, Bailin Hao and H.C. Lee [ PDF]
|
Global Comparison of
Thirteen Bacterial Complete Genomes and Phylogeny A global comparison of the thirteen complete bactrial genomes available from the GenBank to May of 1998 is made by studying genome-genome correlations of distributions of frequency of occurrence of oligonucleotides (6-mers and 7-mers, respectively) tagged by under-represented palindromes (4-mers and 5-mers respectively) that are mostly recognition sites of bacterial restriction enzymes. Whereas frequency distributions of untagged oligonucleotides have little species dependence and correlations of such distributions show no structure, frequency distributions of the tagged oligonucleotides are strongly species dependent, and correlations of such distributions are highly structured and seem to yield useful phylogenetic information. In particular, correlations between species distant on the rRNA phylogenetic tree are observed, several of which corroborate results of recent analyses of gene homology suggesting the frequent occurrence of lateral gene transfer between species. | 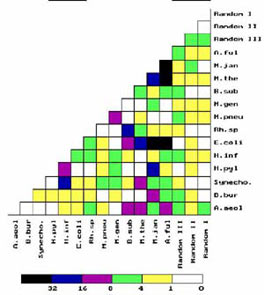 |
The Bio-Cross Project - A life science oriented integrated interdisciplinary research project in natural science, engineering and computer science, H.C. Lee (Proposal in NSC and the Ministry of Education competition for centers of research excellence, October, 2000) (Chinese & English) [PDF]
Report on the NCTS Core Program:
Biology Inspired Theoretical Science (BITS)
H.C. Lee (Report to External Review Committees to National
Center for Theoretical Sciences, March 15-16 and May 11-12, 2000) [PDF]